Connectomics
A connectome refers to the full wiring diagram of the nervous system.
At birth, C. elegans has 218 neurons. 82 new neurons are incorporated before the end of larval development. These include sensory, motor, and interneurons that reside throughout the body, suggesting a system-wide wiring remodeling as the animal matures.
An adult connectome was mapped more than 30 years ago by combining partial reconstruction data of more than five different individuals (The Mind of a Worm, 1986). This landmark study led to the first connectome, serving as the guide for functional interrogation in adults. However, on its own it is insufficient to address how the circuit develops and varies among individuals.
New technologies, from sample preparation to image processing, improve the fidelity and throughput of serial electronic microscopy (EM). With the Samuel and Lichtman groups, we have built a pipeline to reconstruct and compare individual connectomes (Mulcahy et al., 2018). We have completed brain reconstruction from eight isogenic individuals from hatching to adulthood (Witvliet et al., 2021)
Comparing them has told us how the C. elegans brain matures. In brief, we have uncovered the following patterns for brain maturation at the cellular and network levels:
- The overall geometry of the brain is preserved from birth to adulthood. It is upon this constant scaffold that substantial changes in chemical synaptic connectivity emerge.
- There are substantial connectivity differences that make each brain partly unique, among an isogenic population.
- Synaptogenesis strengthens connections that exist at birth and create new connections as animals mature. They maintain central decision-making circuitry whereas sensory and motor pathways substantially remodel.
- Collective changes in the network alter information processing. With age, the brain progressively becomes more feedforward and discernibly modular.
Future work:
We need to extend the study of the development connectomics.
First, we have not included gap junctions, critical components of the nervous system, in our analysis. Improvements in sample preparation and analysis are needed. Second, we have analyzed only one connectome at most time points. Increased throughput and the analysis of many animals at each age will allow analysis of the statistical properties of synaptic connectivity.
Comparative connectomics is needed to understand the origin of similarities and differences in structure and behaviour, within and across species. High-throughput and high-resolution electron microscopy are necessary to establish the foundation for understanding how genes, experience, and evolution create the behaving adult.
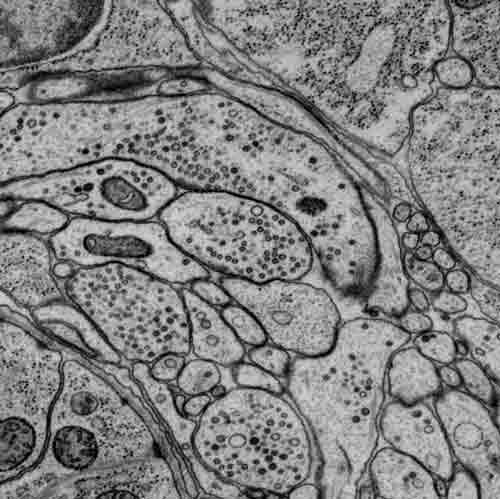
Part of a scanning electron micrograph highlighting synapses in the brain (nerve ring) of a first-stage larva. Imaged by James Mitchell, the Samuel Lab.
Volumetric reconstruction of neurites and muscles of a C. elegans adult brain.
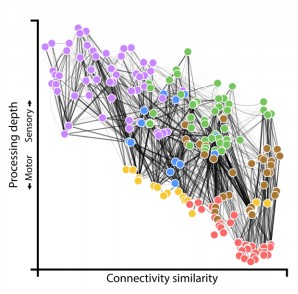
The wiring diagram for the adult connectome with each cell colored by its assigned module (Witvliet et al. 2021)
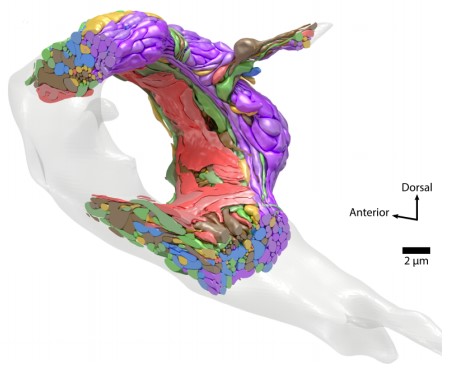
A 3D model of the adult brain with each cell colored by its assigned module (Witvliet et al. 2021). Click here for the video.
Neural Coding of Behaviors
The compact nervous system of C. elegans allows us to address neural codes of behaviors and behavioral plasticity at multiple levels (cell, circuit, and network).
We are interested in following questions:
- How neurons organize into circuits by regulated synapse development.
- How circuits gather information to determine appropriate motor responses.
- How these motor responses adapt, to changes in the body or environments.
Specifically, we identify and document the relationships between both the motor behaviors and the activity pattern of neurons as well as the effect of perturbation of neuronal connections. These correlations form the components of the functional connective of the neural circuit. They are determined by an array of tools – genetics, electrophysiology, real-time calcium imaging, and optogenetics.
Through these studies, we reveal following operational principles at the locomotory circuit:
- Body wall muscles generate L-VGCC (ELG-19)-dependent, Ca-driven action potentials. Trains of action potentials underlie muscle contraction (Gao and Zhen, 2011).
- Excitatory motor neurons in the ventral nerve cord form a chain of intrinsic oscillators that derive locomotor rhythms. The oscillation is driven by P/Q/N-VGCC (UNC-2). These motor neurons are essentially a compressed spinal cord, integrating the role of the command interneuron, motor neuron, and proprioceptive neurons.
- Two classes of motor neurons, the A and B, are oscillators for backward and forward locomotion respectively. A has higher intrinsic activity than B (Gao et al., 2018; Xu et al., 2018; Wen, Gao and Zhen, 2018).
- Descending premotor interneurons regulate the oscillation of excitatory motor neurons to change propensity between forward and backward movement.
- Two separate classes of premotor interneurons regulate the forward and backward oscillators, with mixed synaptic connections (gap junctions and chemical synapses). For reversal, AVA shut A’s activity by gap junctions at rest, and potentiate A by chemical synapse when stimulated (Kawano et al., 2011; Gao et al., 2018; Wen, Gao and Zhen, 2018). For forward, AVB potentiates B’s activity by gap junction (Xu et al., 2018; Wen, Gao and Zhen, 2018).

Optogenetic activation of inhibitory motor neurons by NpHR in the ventral nerve cord leads to relaxation along the body. Yellow indicates light on.
Whole-brain calcium imaging by a GCaMP sensor reveals symmetric activity pattern of the left-right brain.
Molecular Determinants of Excitability
Neurons are excitable cells. They generate changes in membrane potential to encode and transmit information. The resting membrane potential (RMP) of the neuron is a key determinant of its excitability.
Leak channels generate the sodium and potassium currents that shape the RMP. We examine how these leak channels affect the development and activity of individual neurons and the neuronal network. We are examining the physiological roles of leak channels using electrophysiology, imaging, RNA sequencing, and genome editing.
Sodium leak: by screening for genetic mutations that affect C. elegans motor patterns, we discovered the pore (NCA-1, NCA-2), accessory (UNC-79, UNC-80), and ER-delivery subunits (NLF-1) of the sodium leak channel (Yeh et al., 2008; Xie et al., 2013; Gao et al., 2015). We identified and showed the functional conservation of the mouse channel subunit of NLF – FAM155A/mNLF-1 (Xie et al., 2013). We discovered genetic mutations that increase the leak channel activity, from C. elegans (Yeh et al., 2008) to human patients (Aoyagi et al., 2015).
Disease Models
More than 80% of the C. elegans proteomes have human homologs. The ease of genetic manipulation and the collection of experimental tools to examine the anatomy and activity of neurons and neuronal connections allow for the system to model neurological disorders and address their defects across the synapse, cell, and circuit resolution. We have established the C. elegans models for ALS (Murakami et al., 2015), CLIFAHDD (Aoyagi et al., 2015), and JBS (Chitturi et al. 2018). In all models, locomotory behaviours are the functional readout to dissect the underlying molecular, cellular, and circuit mechanisms of the causative genetic mutations.
Through our work on neuronal excitability, we identified the first case of CLIFAHDD, a neurodevelopmental disorder caused by gain-of-function mutations in the sodium leak channel (Aoyagi et al., 2015). We have now established the mouse model for CLIFAHDD. An ongoing project in our lab is to further our understanding of the circuit mechanism that underlies this disorder.